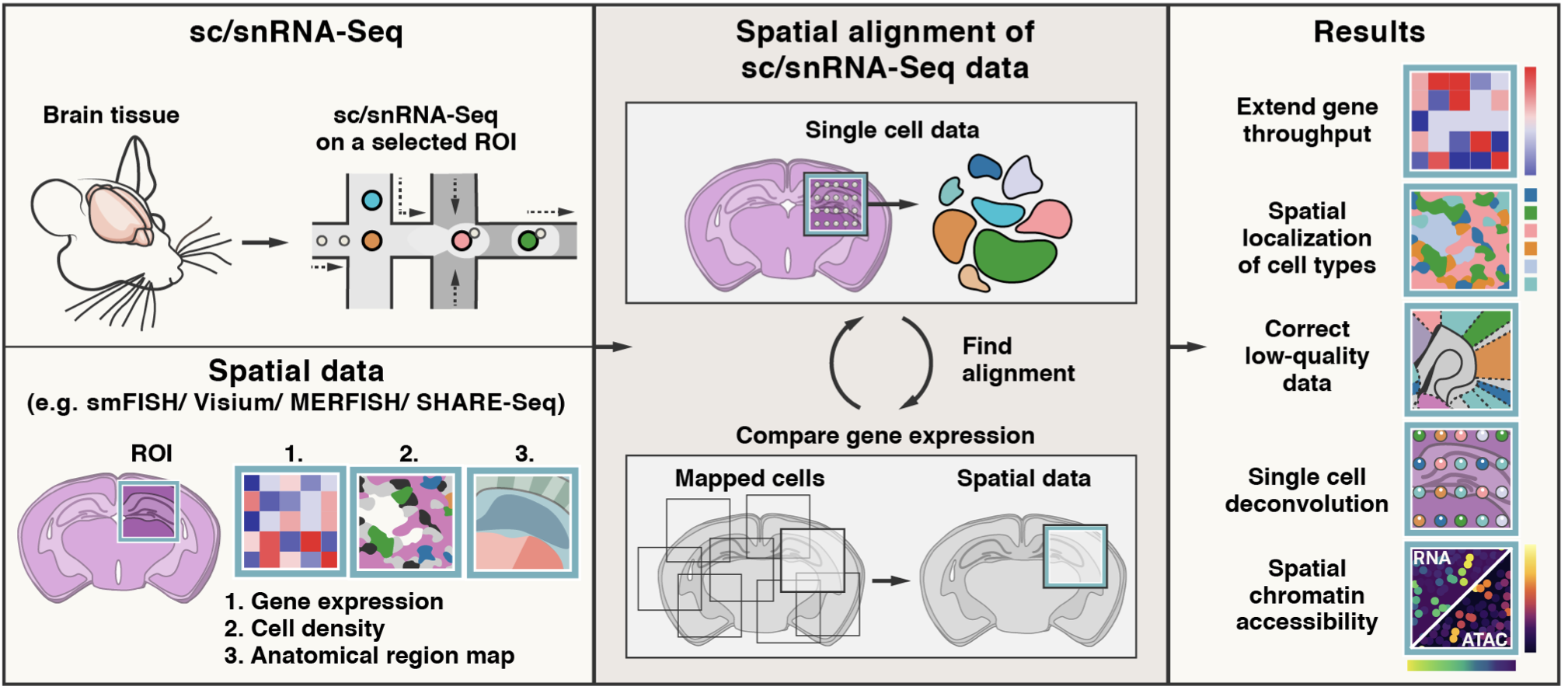
TANGRAM
Tangram is a Python package, written in PyTorch and based on scanpy , for mapping single-cell (or single-nucleus) gene expression data onto spatial gene expression data. The single-cell dataset and the spatial dataset should be collected from the same anatomical region/tissue type, ideally from a biological replicate, and need to share a set of genes. Tangram aligns the single-cell data in space by fitting gene expression on the shared genes. The best way to familiarize yourself with Tangram is to check out our tutorials.
Tangram News
On Jan 28th 2021, Sten Linnarsson gave a talk at the WWNDev Forum and demostrated their mappings of the developmental mouse brain using Tangram.
On Mar 9th 2021, Nicholas Eagles wrote a blog post about applying Tangram on Visium data.
The Tangram method has been used by our colleagues at Harvard and Broad Institute, to map cell types for the developmental mouse brain -see Fig. 2 [`Nature(2021)<https://www.nature.com/articles/s41586-021-03670-5>`_]
Tangram is now officially a part of Squidpy
Citing Tangram
Tangram has been released in the following publication
Biancalani* T., Scalia* G. et al. - _Deep learning and alignment of spatially-resolved whole transcriptomes of single cells in the mouse brain with Tangram biorXiv 10.1101/2020.08.29.272831 (2020)
Release Notes
1.0.0 2021-08-06 - Initial Release


